Why is genetic verification the best methodology for antibody validation?
- Brandon Fan

- Apr 10, 2020
- 4 min read
Updated: Apr 13, 2020
In my last blog (Blog#3), I compared the five approaches (pillars) for antibody validation, i.e. orthogonal approach, independent antibody approach, recombinant expression approach, captured mass spectrometry approach, and genetic approach (Uhlen et al., 2016). In this blog, I will focus on genetic verification and explain why this approach is the best method for antibody validation.
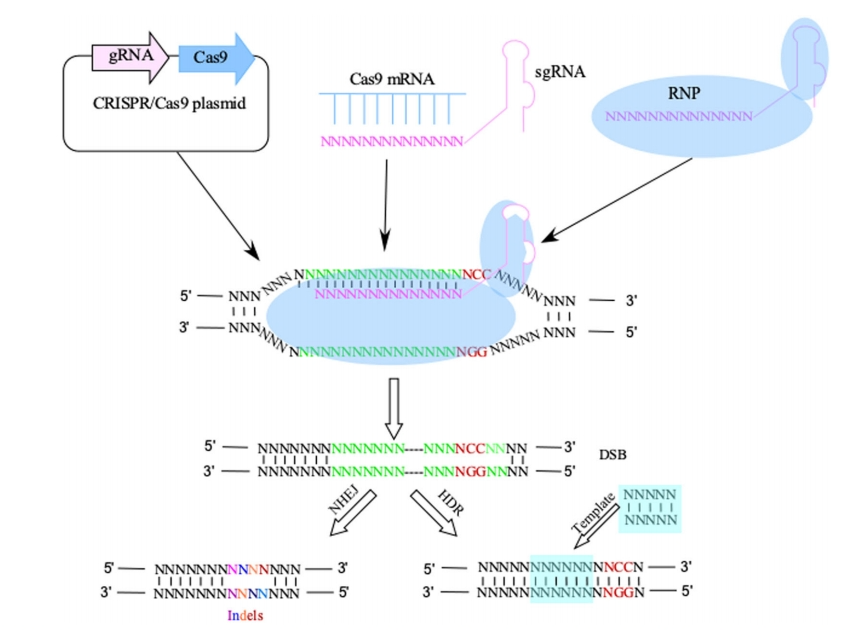
Over the past two decades, short RNA-based silencing technologies (also referred to as RNA interference or RNAi) have rapidly evolved and been widely used in basic research and therapeutic applications (Dykxhoorn et al., 2003; Fischer, 2015). In parallel, CRISPR-associated RNA-guided endonuclease Cas9 (CRISPR-Cas9) has revolutionized genome editing and advanced the field to make genomic disruptions much simpler and more scalable than we could have ever imagined (Hatada and Horii, 2017; Hsu et al., 2014). The traditional methods of gene knockout technology such as homologous recombination alone, or in combination with CRISPR-Cas9 (Bellin, 2018), are still being used with the modern technologies due to their proven track record of success. In recent years, these gene manipulation technologies have been increasingly used to tackle the thorny issues in the field of research antibodies: lack of specificity, irreproducibility, batch-to-batch variations, and incorrect applications (Baker, 2015; Berglund et al., 2008; Bradbury and Pluckthun, 2015), just to name a few.
To answer the question of why genetic verification is the best approach to validate antibody specificity, we have to examine the definition of specificity. Specificity refers to an antibody’s ability to bind to (have affinity) and only to (being selective) its target antigen (Pillai-Kastoori et al., 2020). In other words, an antibody should recognize and bind to its target protein, but it should not bind to other proteins. This exclusivity demands high-standard validating techniques, among which genetic verification is the best choice. First, techniques other than genetic verification are an indirect validation, whereas the genetic strategy is a direct validation. Knocking down or knocking out a gene reduces or eliminates its encoded protein, thereby generating a true negative control for the wild-type positive control (Pillai-Kastoori et al., 2020). A lack of the antibody signal in genetically altered cells, detected through enhanced chemiluminescence (ECL) or fluorescent light emission, coupled with the positive signal in wild-type cells, demonstrates that the antibody specifically and selectively binds to the target protein and no other proteins. In contrast, a positive signal from genetically altered cells suggests that the antibody has off-target or nonspecific binding. Second, genetic verification can be performed in multiple applications. For example, once a gene is knocked down or knocked out, cell lysates can be used for antibody validation in Western blotting, (co-)immunoprecipitation, and chromatin immunoprecipitation. Alternatively, genetically altered cells can be fixed and subjected to immunocytochemical or flow cytometric analyses. This cross-examination of antibody specificity provides another layer of assurance that the tested antibody is specific for its intended target.
Of course, genetic verification is not without shortcomings. One of the shortcomings is that because certain genes are so essential for cell survival, the knock out of these genes will be lethal. In this regard, gene silencing can circumvent this disadvantage by decreasing, rather than eliminating, the expression of the target protein (Moore et al., 2010). Using small interfering RNA (siRNA) to knock down genes is widely used in many laboratory setting because it is cost-effective and less time consuming. Nonetheless, the knockdown by siRNA is transient (Moore et al., 2010), and it is not applicable for long-half-life proteins as these proteins can remain in the cells for a prolonged period of time although its mRNA has been knocked down by siRNA. In this regard, short hairpin RNA (shRNA) has obvious advantages in that 1) lentiviral shRNA can be stably integrated into a cell’s genome and result in a more complete knockdown, 2) it is particularly powerful for difficult-to-target and nondividing cells (Lambeth and Smith, 2013).
At GenuIN Biotech LLC, we have developed a proprietary shRNA-mediated gene knockdown and knockout platform to test antibody specificity. We are here to safeguard the quality of antibodies you use so that your results are reproducible, and your conclusions are sound. In this sense, we are here to help you save time so that you can focus on more important things in your research. Please check our website for antibodies and/or reagents and let us know if you need further information or assistance.
References
Baker, M. (2015). Reproducibility crisis: Blame it on the antibodies. Nature 521, 274-276.
Bellin, M. (2018). Crispr/Cas9 homologous recombination (HR). Drug Discov Today Technol 28, 1-2.
Berglund, L., Bjorling, E., Oksvold, P., Fagerberg, L., Asplund, A., Szigyarto, C.A., Persson, A., Ottosson, J., Wernerus, H., Nilsson, P., et al. (2008). A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7, 2019-2027.
Bradbury, A., and Pluckthun, A. (2015). Reproducibility: Standardize antibodies used in research. Nature 518, 27-29.
Dykxhoorn, D.M., Novina, C.D., and Sharp, P.A. (2003). Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4, 457-467.
Fischer, S.E.J. (2015). RNA Interference and MicroRNA-Mediated Silencing. Curr Protoc Mol Biol 112, 26 21 21-26 21 25.
Hatada, I., and Horii, T. (2017). CRISPR/Cas9. Methods Mol Biol 1630, 37-42.
Hsu, P.D., Lander, E.S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262-1278.
Lambeth, L.S., and Smith, C.A. (2013). Short hairpin RNA-mediated gene silencing. Methods Mol Biol 942, 205-232.
Moore, C.B., Guthrie, E.H., Huang, M.T., and Taxman, D.J. (2010). Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol 629, 141-158.
Pillai-Kastoori, L., Heaton, S., Shiflett, S.D., Roberts, A.C., Solache, A., and Schutz-Geschwender, A.R. (2020). Antibody validation for Western blot: By the user, for the user. J Biol Chem 295, 926-939.
Uhlen, M., Bandrowski, A., Carr, S., Edwards, A., Ellenberg, J., Lundberg, E., Rimm, D.L., Rodriguez, H., Hiltke, T., Snyder, M., et al. (2016). A proposal for validation of antibodies. Nat Methods 13, 823-827.


Comments